Alysha De Livera, PhD
Senior Lecturer in Statistics
Department of Mathematics and Statistics, La Trobe University, Melbourne, Australia
Dr De Livera is an academic in Statistics in the Department of Mathematics and Statistics, the Director of the Statistical Consultancy Platform, and the Partnership Coordinator for Mathematical and Physical Sciences at La Trobe University. She enjoys both research and lecturing in an interactive environment.
Alysha works closely with scientific investigators from a diverse range of backgrounds on multidisciplinary research projects, contributing her statistical expertise to a wide range of problems in medicine, public health, epidemiology, and biology. Her research on statistical methods and software is motivated by issues that arise from these studies.
Alysha’s methodological research interests in Statistics include meta-analysis, statistical methods for high-dimensional data analysis, and methods for handling missing values. In her recent work, Alysha has led the development of novel statistical methods and software for the analysis of high-dimensional biological data including metabolomics. Her methodological work has been published in top-tier journals including the Journal of the American Statistical Association, Bioinformatics, Analytical Chemistry and Nucleic Acids Research, and she also serves as a regular reviewer for many scientific journals.
Alysha actively engages in mentoring, consulting, and supervision of the students, and co-ordinates multiple statistical subjects in the Department of Mathematics and Statistics. Alysha is a former co-chair of the Biostatistics and Bioinformatics Section of the Statistical Society of Australia (SSA), helps promote the discipline of Statistics and Data Science in Australia, and advocates women in STEM.
Affiliations
Research
Featured Publications
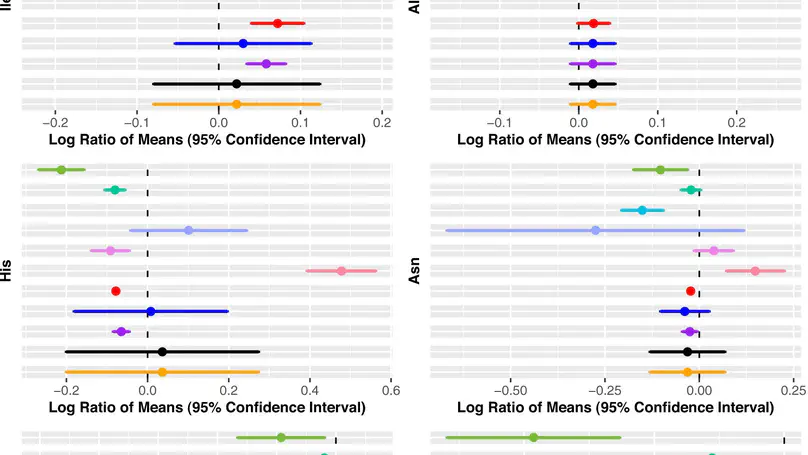
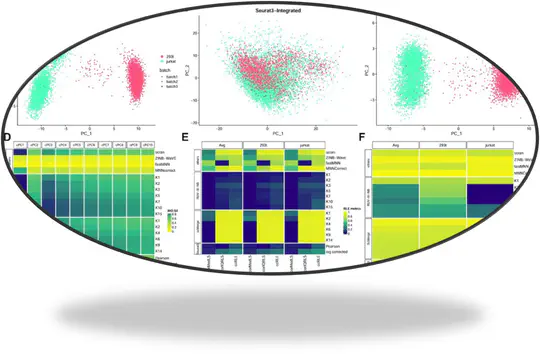
Motivation- Meta-analysis methods widely used for combining metabolomics data do not account for correlation between metabolites or missing values. Within- and between-study variability are also often overlooked. These can give results with inferior statistical properties, leading to misidentification of biomarkers. Results- We propose a multivariate meta-analysis model for high-dimensional metabolomics data (MetaHD), which accommodates the correlation between metabolites, within- and between-study variances, and missing values. MetaHD can be used for integrating and collectively analysing individual-level metabolomics data generated from multiple studies as well as for combining summary estimates. We show that MetaHD leads to lower root mean square error compared to the existing approaches. Furthermore, we demonstrate that MetaHD, which exploits the borrowing strength between metabolites, could be particularly useful in the presence of missing data compared with univariate meta-analysis methods, which can return biased estimates in the presence of data missing at random. Availability and implementation- The MetaHD R package can be downloaded through Comprehensive R Archive Network (CRAN) repository. A detailed vignette with example datasets and code to prepare data and analyses are available.
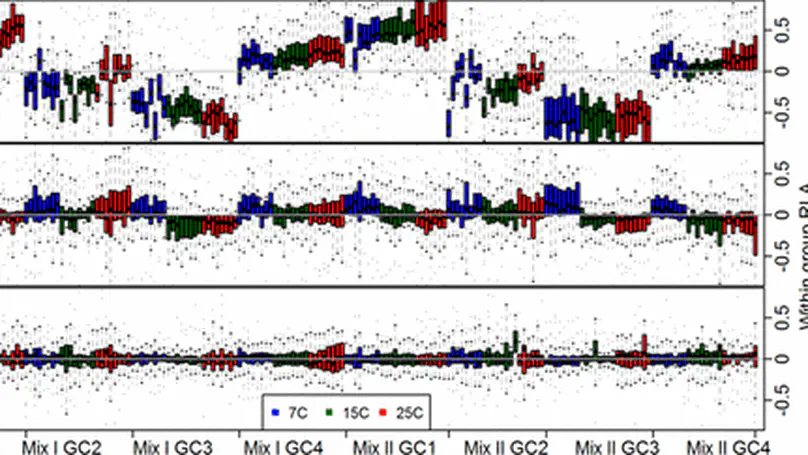
Metabolomics experiments are inevitably subject to a component of unwanted variation, due to factors such as batch effects, long runs of samples, and confounding biological variation. Although the removal of this unwanted variation is a vital step in the analysis of metabolomics data, it is considered a gray area in which there is a recognized need to develop a better understanding of the procedures and statistical methods required to achieve statistically relevant optimal biological outcomes. In this paper, we discuss the causes of unwanted variation in metabolomics experiments, review commonly used metabolomics approaches for handling this unwanted variation, and present a statistical approach for the removal of unwanted variation to obtain normalized metabolomics data. The advantages and performance of the approach relative to several widely used metabolomics normalization approaches are illustrated through two metabolomics studies, and recommendations are provided for choosing and assessing the most suitable normalization method for a given metabolomics experiment. Software for the approach is made freely available.
Contact
- a.delivera@latrobe.edu.au
- +61 3 9479 6587
- Melbourne, Australia 3000
- DM
- DM