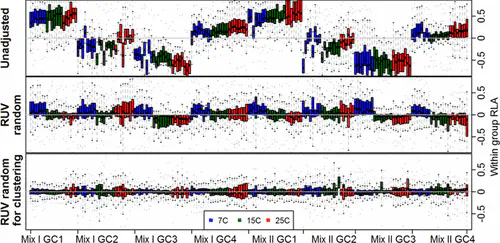
Abstract
Metabolomics experiments are inevitably subject to a component of unwanted variation, due to factors such as batch effects, long runs of samples, and confounding biological variation. Although the removal of this unwanted variation is a vital step in the analysis of metabolomics data, it is considered a gray area in which there is a recognized need to develop a better understanding of the procedures and statistical methods required to achieve statistically relevant optimal biological outcomes. In this paper, we discuss the causes of unwanted variation in metabolomics experiments, review commonly used metabolomics approaches for handling this unwanted variation, and present a statistical approach for the removal of unwanted variation to obtain normalized metabolomics data. The advantages and performance of the approach relative to several widely used metabolomics normalization approaches are illustrated through two metabolomics studies, and recommendations are provided for choosing and assessing the most suitable normalization method for a given metabolomics experiment. Software for the approach is made freely available.